Big Data Knowledge Graph – Knowledge Graph + flask based big data NLP medical knowledge Q&A system (the most detailed explanation and source code on the net / recommended collection)
-
- I. Project overview
- Second, the basic process of medical knowledge Q&A system that realizes knowledge graphs
- III. Version numbers used for project tools
- IV. Installation and use of required software
- V. System implementation
- VI. Building the neo4j knowledge graph
- VII. Knowledge map data entry
- VIII. Cypher-based queries
- IX. Project initiation realization methodology and steps
- X. Using Python to realize the addition, deletion, modification, data query and maintenance
- XI. Conclusion
I. Project overview
A knowledge graph is a network formed by connecting knowledge together. It consists of nodes and edges, nodes are entities, edges are relationships between two entities, and both nodes and edges can have attributes. In addition to querying the attributes of an entity, the knowledge graph can easily find related entity and attribute information from an entity by traversing the relationship.
KBQA medical Q&A system based on knowledge graph + flask is based on the medical aspects of knowledge Q&A, by building a knowledge graph in the medical field, and with this knowledge graph to complete the automatic Q&A and analysis services. KBQA medical Q&A system based on knowledge graph+flask uses neo4j as a storage, the maximum forward matching used for knowledge graph modeling in this system is a greedy algorithm, which starts matching from the beginning of the sentence and selects the longest word each time. It is relatively fast in terms of speed as it requires only one traversal. The algorithm is relatively simple, easy to implement and understand, and does not require complex data structures. For the case where most of the Chinese text is left-oriented, max-forward matching is usually able to slice it better. Contrary to Maximum Forward Matching, Maximum Backward Matching starts matching from the end of the sentence and selects the longest word each time. It is suitable for most right-oriented Chinese text. Bidirectional max-matching combines the advantages of max-forward and max-backward matching by matching in both directions and then selecting the one with the smaller number of clauses. This method integrates the leftward and rightward features and improves the accuracy of the syncopation to perform a cypher query with the keywords and return the corresponding result query statement as a Q&A. Later I designed a simple Flask based chatbot application that utilizes nlp natural language processing to return results based on the user’s questions through a medical AI assistant, where both the user inputs and the outputs returned by the system are automatically stored together in a sql database.
In terms of multi pattern matching, the Aho-Corasick algorithm is specialized for searching multiple patterns (keywords) in a text simultaneously. Compared to the violent search algorithm, the Aho-Corasick algorithm has a lower time complexity and performs more significantly in this Knowledge Graph modeling question and answer system. In terms of linear time complexity, the stage of performing preprocessing, the Aho-Corasick algorithm constructs a deterministic finite automaton (DFA) such that the time complexity in the search stage is O(n), where n is the length of the text to be searched. This linear time complexity makes the algorithm very efficient in this application. In terms of flexibility, the Aho-Corasick algorithm can easily add and remove pattern strings during the construction of the finite automaton without reconstructing the entire data structure, improving the flexibility and maintainability of the algorithm.
Second, the basic process of medical knowledge Q&A system that realizes knowledge graphs
-
Configure the required environment (jdk,neo4j,pycharm,python, etc.)
-
Crawl the required medical data to get the basic medical data needed.
-
Data cleansing process for medical data.
-
on the basis ofgreedy algorithmPerforms a split-word strategy.
-
Relationship Extraction Definition and Entity Identification.
-
Knowledge graph modeling.
-
on the basis of
Aho-CorasickAlgorithm for multi-pattern matching. -
Design a Flask-based chatbot AI assistant.
-
Design user input and system output record data automatically stored to sql database.
III. Version numbers used for project tools
Neo4j version: Neo4j Desktop 1.4.15.
neo4j inside medical system database version: 4.4.5.
Pycharm version: 2021.
JDK version: jdk1.8.0_211.
MongoDB version: MongoDB-windows-x86_64-5.0.14.
flask version: 3.0.0
IV. Installation and use of required software
(i) Installation of JAVA
1. Download the java installer:
Official website download link:https://www.oracle.com/java/technologies/javase-downloads.html
I downloaded the version of the JDK-1.8, JDK version of the choice must be appropriate, the version is too high or too low may lead to subsequent neo4j can not be used.
After installing the JDK you have to start configuring environment variables. The steps to configure environment variables are as follows:
Right-click this computer – click Properties – click Advanced System Settings – click Environment Variables
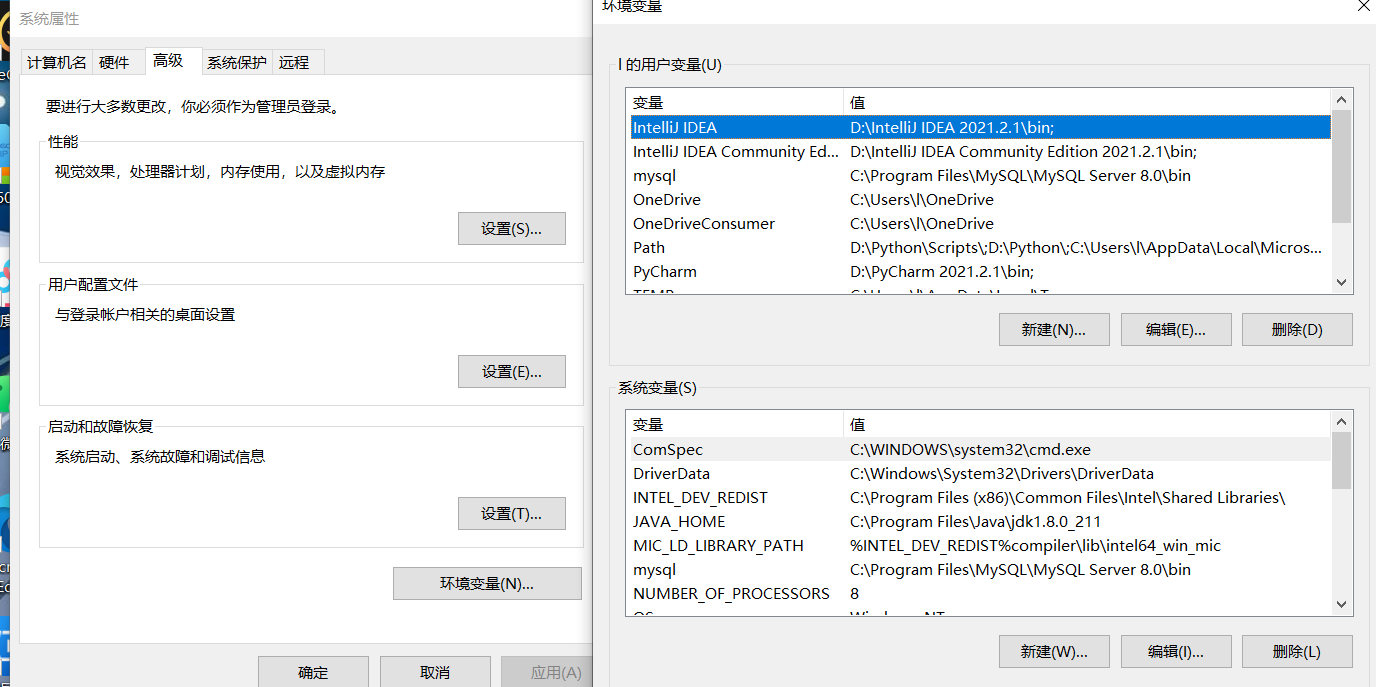
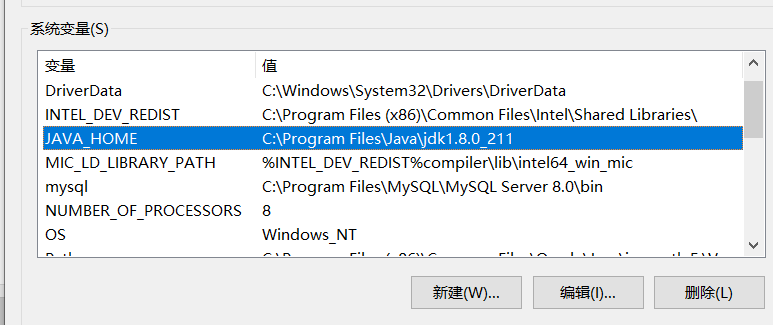
In the following system variables area, create a new environment variable, named JAVA_HOME, variable value set to just JAVA installation path, I here is C:\Program Files\Java\jdk1.8.0_211
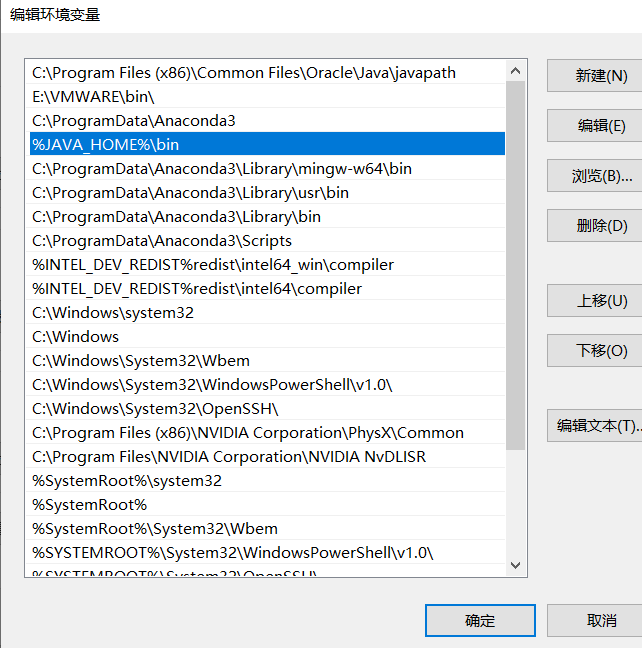
Edit the Path in the System Variables section, click New, and type %JAVA_HOME%\bin
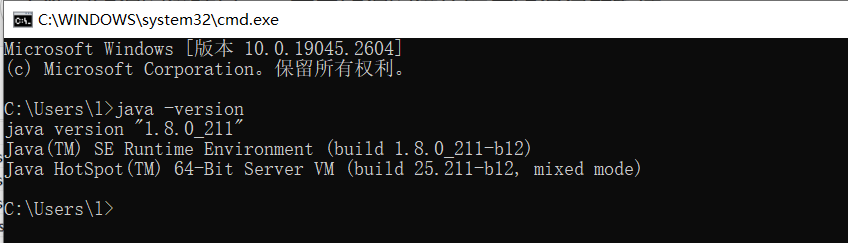
Open the command prompt CMD (WIN + R, enter cmd), enter java -version, if prompted Java version information, it proves that the environment variable configuration is successful.
2. After installing the JDK, you can install the neo4j
2.1 Download neo4j
Official download link:https://neo4j.com/download-center/#community
Or you can just download the link I uploaded to my cloud drive:
Neo4j Desktop Setup 1.4.15.exe
Link:https://pan.baidu.com/s/1eXw0QjqQ9nfpXwR09zLQHA?pwd=2023
Extraction code: 2023
After opening the default path, you can set your own computer according to their own settings, and then wait for the start on the line!
When we open it, we create a new database called: “Q&A system based on the medical field”.
For more information, please see the picture below.
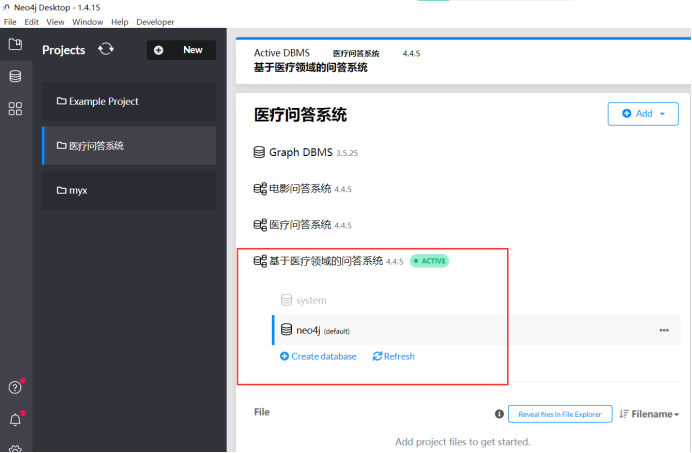
The database used is version 4.4.5, and other database parameter information is listed below:

V. Overall catalog of project structures
├── pycache \\\ Compilation result save directory
│ │ ├── answer_search.cpython-39.pyc
│ │ ├── question_classifier.cpython-39.pyc
│ │ │── Question-based parsing.cpython-39.pyc
│ │ │── Query based on question parsing results.cpython-39.pyc
│ │ │── Problem-based classification.cpython-39.pyc
├── data\\ of this project
│ └── medical.json \ \ The data of this project, through build_medicalgraph.py import neo4j
├── dict
│ ├── check.txt \ diagnostic check item entity library
│ ├── deny.txt \ Negative words thesaurus
│ ├── department.txt \ medical subject entity library
│ ├── disease.txt \ Disease Entity Library
│ ├── drug-txt \ drug physical library
│ ├── food.txt \ Food Entity Library
│ ├── producer.txt \ Pharmacy for sale
│ └── symptom.txt \ Disease Symptom Entity Library
├── prepare_data \crawler and data processing
│ ├──__pycache__
│ ├── build_data.py \ database operation scripts
│ ├── data_spider.py \ Network Information Collection Scripts
│ └── max_cut.py \ Dictionary-based max forward/backward scripts
│ ├── Processed medical data.xlsx
│ ├── MongoDB data to json format data file.py
│ ├── Medic.csv exported from MongoDB
│ ├──data.json #Data exported from MongoDB in json format
├── static \static resource files
├── templates
│ ├──index.html \ \ Q&A system front-end UI page
├── app.py \start flask quiz AI script main program
├── question_classifier.py \ Question Type Classifier Script
├── answer_search.py \based question-response scripts
├── build_medicalgraph.py \ Build Medical Graph Scripts
├── chatbot_graph.py \ medical AI assistant Q&A system bot script
├── question_parser.py [\based question parsing script](file://based question parsing script)
├── Delete all relationship.py \ optional script files
├── Delete relationshipchain.py \ \ Disposable script file
├── Two nodes newly added relationship.py \ \ optional script file
├── Interaction Match all nodes.py \ \ optional script file
Here the chatbot_graph.py script starts its analysis with the chatbot_graph.py file that needs to be run.
The script constructs a question and answer class, ChatBotGraph, defining a member variable classifier of type QuestionClassifier, a member variable parser of type QuestionPase, and a member variable searcher of type AnswerSearcher.
The question_classifier.py script constructs a class QuestionClassifier for question classification, defining member variables such as feature word paths, feature words, domain actree, lexicon, and question words for question sentences. question_parser.py question sentences need to be parsed after question classification. The script creates a QuestionPaser class that contains three member functions.
After parsing the answer_search.py question you need to query the parsed results. The script creates an AnswerSearcher class. Similar to build_medicalgraph.py, this class defines the Graph class member variable g and returns the maximum number of answers enumerated, num_list.The class has two member functions, a query principal and a reply module.
The framework of the question and answer system is built with scripts such as chatbot_graph.py, answer_search.py, question_classifier.py, and question_parser.py.
V. System implementation
Data Capture and Storage
Install the MongoDB database:
MongoDB official download address:
https://www.mongodb.com/try
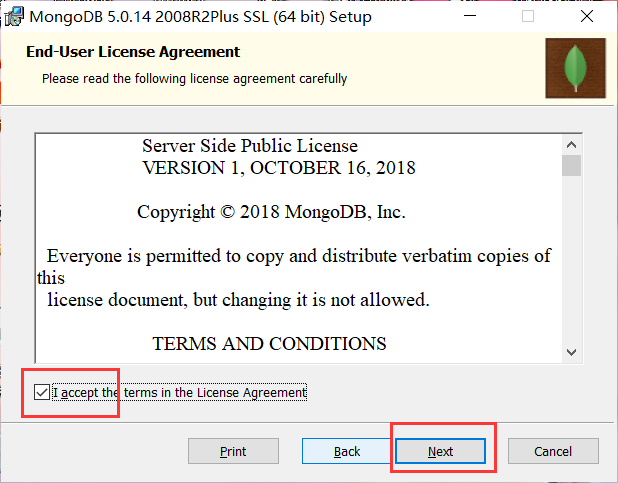
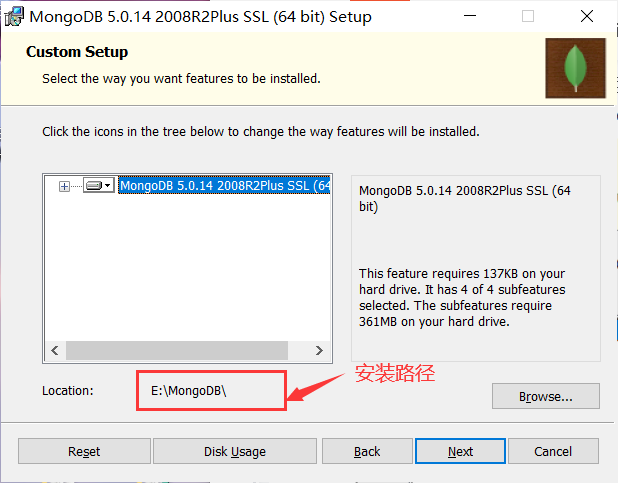
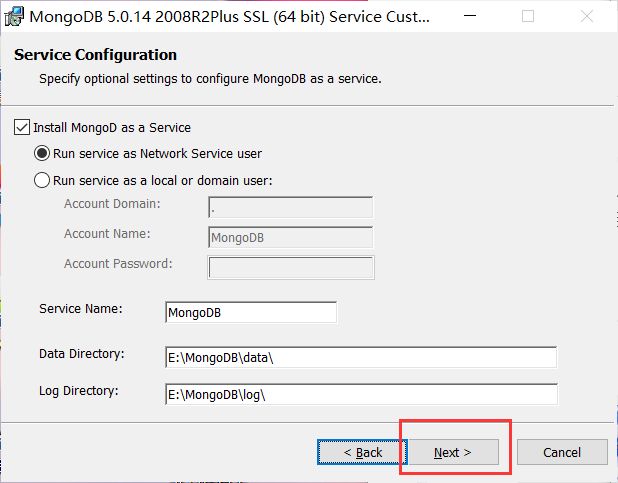
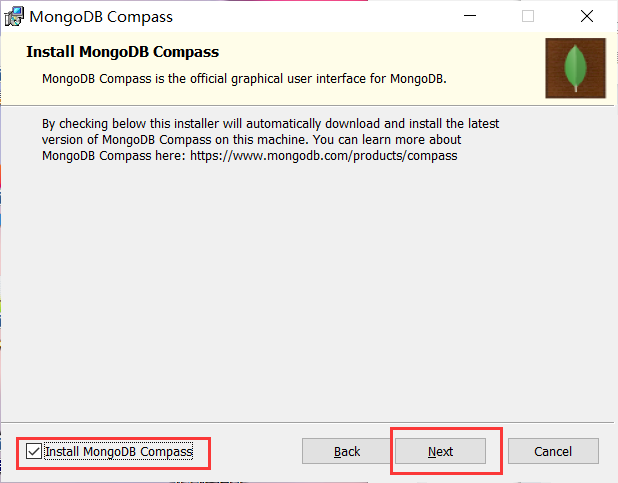
Installation complete:
Then we do the MongoDB environment configuration:
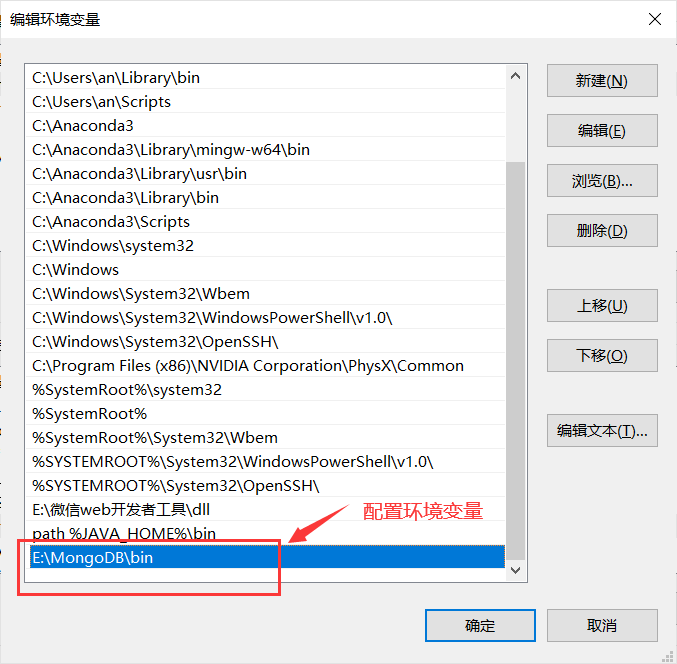
Add E:\MongoDB\bin to the variable Path
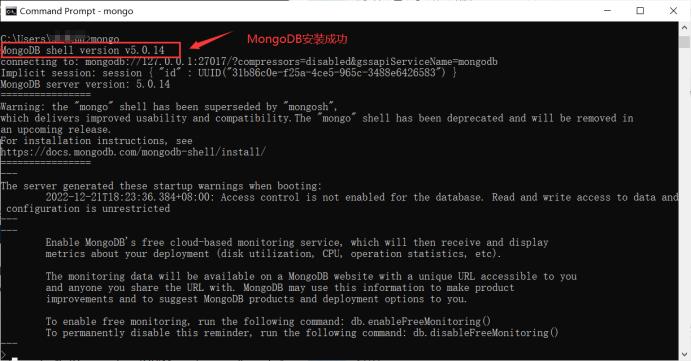
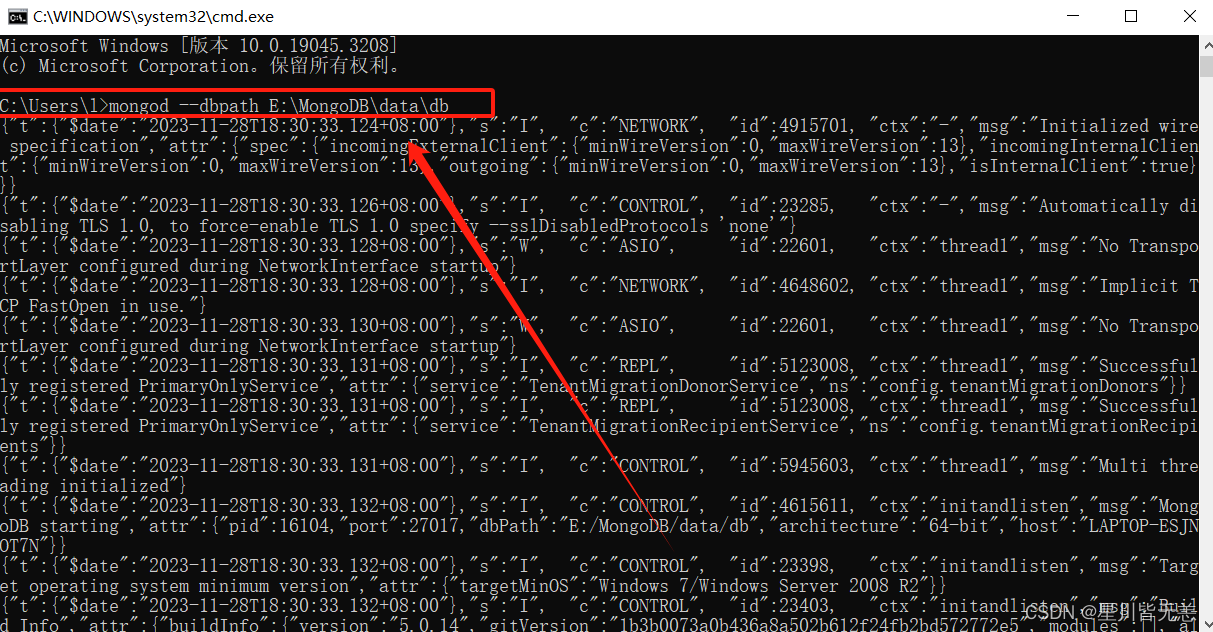
Open a terminal (cmd) and type mongod –dbpath E:\MongoDB\data\db
Type in your browser
127.0.0.1:27017See if the MongoDB service started successfully:
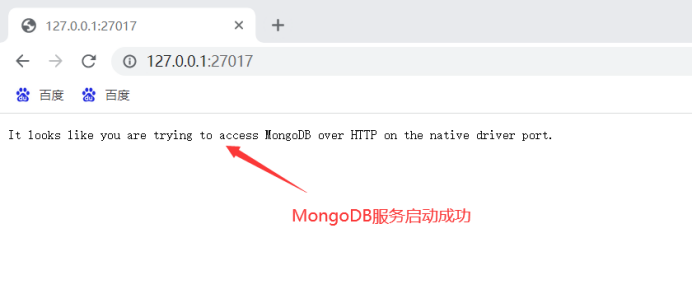
After installing MongoDB start crawling the data:
The data was sourced from medical-seeking.com:http://jib.xywy.com/
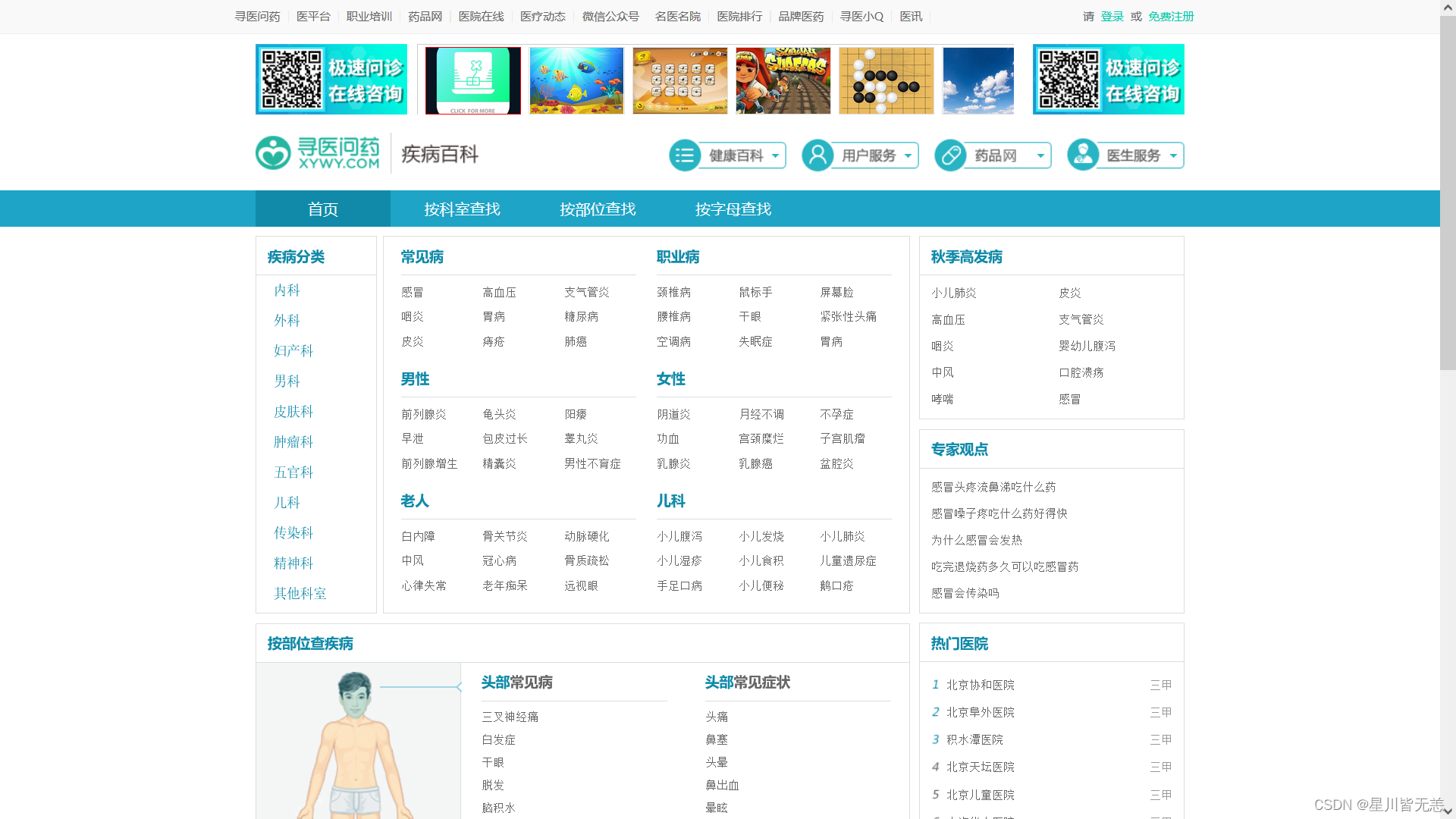
Specific disease detail pages are listed below:
The links to diseases on the URL were first analyzed, using the cold as an example:
Cold and Flu Links:http://jib.xywy.com/il_sii_38.htm
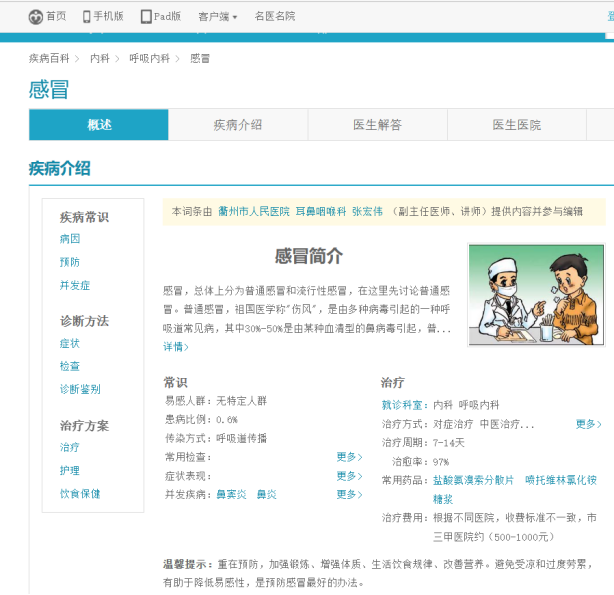
As you can see, it contains the content of the disease profile, causes, prevention, symptoms, tests, treatment, complications, diet and health care details page. Here we want to use a crawler to collect the information. To collect the corresponding data under the url, the specific crawler code is as follows:
Previously, the old version of insert was deprecated and would give warnings again. insert has been replaced with insert_one so that you no longer get warnings about the insert method being deprecated.
'''Healthcare data collection based on doctor search''''
# Use the insert_one or insert_many methods. Provides more flexibility and supports additional features such as the _id value of the document returned after insertion.
class MedicalSpider:
def __init__(self):
self.conn = pymongo.MongoClient()
self.db = self.conn['medical2']
self.col = self.db['data']
def insert_data(self, data):
# Insert a single document using the insert_one method
self.col.insert_one(data)According to the url, request html
A function to get the HTML content of a given URL, using Python’s requests library. In this code, you set the request’s headers (User-Agent), proxies (proxies), and then use therequests.get method gets the page content and specifies the encoding as ‘gbk’.
'''Based on url, request html'''
def get_html(self,url):
headers = {
'user-agent': 'Mozilla/5.0 (Windows NT 10.0; WOW64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/69.0.3497.100 Safari/537.36'
}
proxies = {
'http': None,
'https': None
}
html = requests.get(url=url, headers=headers, proxies=proxies)
html.encoding = 'gbk'
return html.textCrawl medical-related information, including disease descriptions, disease prevention, disease symptoms, treatments, and more.
url_parser function (math.)
def url_parser(self, content):
selector = etree.HTML(content)
urls = ['http://www.anliguan.com' + i for i in selector.xpath('//h2[@class="item-title"]/a/@href')]
return urlsurl_parser function function is to parse the incoming HTML content, extract the page disease-related links. Specifically:
etree.HTML(content): Use the etree module from the lxml library to convert HTML content into XPath parsable objects.selector.xpath('//h2[@class="item-title"]/a/@href'): Use the XPath selector to extract all<h2>tabclassattribute"item-title"child node of<a>(used form a nominal expression)hrefattribute to get a set of relative links.['http://www.anliguan.com' + i for i in ...]: Converts relative links into full links, stitched together in the'http://www.anliguan.com'Front.
The role of this function is to parse the incoming HTML content and extract the links related to the disease in the page. Specifically:
The spider_main function is the main crawler logic, looping through the page, crawling different types of medical information and storing the results in the database
''''url analysis'''''
def url_parser(self,content):
selector=etree.HTML(content)
urls=['http://www.anliguan.com'+i for i in selector.xpath('//h2[@class="item-title"]/a/@href')]
return urls
'''Links to major crawls'''
def spider_main(self):
# Collection page
for page in range(1,11000):
try:
basic_url='http://jib.xywy.com/il_sii/gaishu/%s.htm'%page # Disease Description
cause_url='http://jib.xywy.com/il_sii/cause/%s.htm'%page # Disease Causes
prevent_url='http://jib.xywy.com/il_sii/prevent/%s.htm'%page # Disease prevention
symptom_url='http://jib.xywy.com/il_sii/symptom/%s.htm'%page # Disease symptoms
inspect_url='http://jib.xywy.com/il_sii/inspect/%s.htm'%page # Disease check
treat_url='http://jib.xywy.com/il_sii/treat/%s.htm'%page # Disease Treatment
food_url = 'http://jib.xywy.com/il_sii/food/%s.htm' % page # Diet Therapy
drug_url = 'http://jib.xywy.com/il_sii/drug/%s.htm' % page # drugbasic_url、symptom_url、food_url、drug_urlrespond in singingcause_urlbased onpageDifferent URL addresses constructed by variables to access and crawl web pages with different types of information.
–datais a dictionary for storing data retrieved from different URLs.
2. Crawling and information extraction for different URLs:
–data['basic_info'] = self.basicinfo_spider(basic_url)</
